%load_ext autoreload
Probabilistic Principal Component Analysis¶
import os
import time
import torch
import numpy as np
from joblib import dump, load
from sklearn.decomposition import PCA
from pdmtut.core import GenerativeModel
from regilib.core.dynamics.dynamical_state import DynamicalState
store_results = True
load_models = True
Introduction¶
Implementation¶
class PPCAModel(GenerativeModel):
def __init__(self, n_components):
self.n=n_components
self.pca = PCA(n_components=self.n, svd_solver='full', whiten=True)
@property
def n_components(self):
return self.pca.n_components_
def fit_model(self, X, path):
start_time = time.time()
self.pca.fit(X)
elapsed_time = time.time() - start_time
if path is not None:
with open(os.path.join(path, 'training_time.txt'), 'w') as f:
f.write(str(elapsed_time))
def encode(self, X):
# based on https://tvml.github.io/ml1718/slides/probabilistic_pca.pdf
_X = (X - self.pca.mean_).numpy()
W = self.pca.components_ * np.sqrt(
self.pca.explained_variance_[:, np.newaxis])
sigma2 = self.pca.noise_variance_
M = W @ W.T + sigma2*np.eye(self.pca.n_components_)
z = _X @ (W.T @ np.linalg.inv(M).T)
return torch.from_numpy(z)
def decode(self, X):
return torch.from_numpy(self.pca.inverse_transform(X))
def save(self, path):
dump(self, os.path.join(path, 'model.joblib'))
def load(path):
return load(os.path.join(path, 'model.joblib'))
def save_exists(path):
return os.path.isfile(os.path.join(path, 'model.joblib'))
def log_likelihood(self, X):
return torch.from_numpy(self.pca.score_samples(X))
def sample_posterior(self, n_samples, include_noise=False):
z_samples = np.random.normal(0, 1, [n_samples, self.pca.n_components_])
if include_noise:
# x ∼ N(W^Tz, σI)
# TODO: do this more efficiently
samples = np.concatenate(
[np.random.multivariate_normal(
self.pca.inverse_transform(z),
self.pca.noise_variance_ * np.eye(self.pca.n_features_) , 1)
for z in z_samples]
)
else:
# x = W^Tz
samples = self.pca.inverse_transform(z_samples)
return torch.from_numpy(samples)
def __str__(self):
return 'ppca_{}_components'.format(self.n)
Experiment 1a: swiss roll (2-components)¶
import pyvista as pv
from pdmtut.datasets import SwissRoll
pv.set_plot_theme("document")
model_save_path = '../results/swiss_roll/ppca_2_components'
if store_results:
result_save_path = '../results/swiss_roll/ppca_2_components'
pv.set_jupyter_backend('None')
else:
pv.set_jupyter_backend('pythreejs')
result_save_path = None
dataset = SwissRoll(n_samples=100**2, seed=11)
if load_models and PPCAModel.save_exists(model_save_path):
model = PPCAModel.load(model_save_path)
else:
model = PPCAModel(n_components=2)
model.fit_model(dataset.X, result_save_path)
if store_results:
model.save(model_save_path)
Input Representation¶
from pdmtut.vis import plot_representation
z = model.encode(dataset.X)
z_extremes = model.encode(dataset.y_extremes)
z_extremes = torch.cat([z_extremes, z_extremes[[1,2]]])
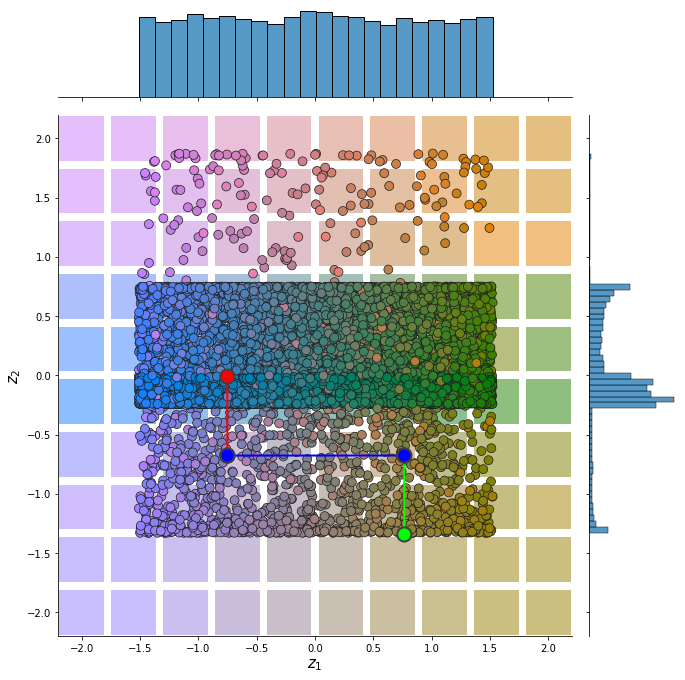
plot_representation(z.numpy(), index_colors=dataset.index_colors, z_extremes=z_extremes, interpolate_background=True, root=result_save_path)
Input Reconstruction¶
from pdmtut.vis import plot_reconstruction
z = model.encode(dataset.X)
x = model.decode(z)
mse = (dataset.unnormalise_scale(dataset.X) - dataset.unnormalise_scale(x)).pow(2).sum(-1).mean()
if result_save_path is not None:
with open(os.path.join(result_save_path, 'reconstruction.txt'), 'w') as f:
f.write(str(mse.item()))
mse
tensor(10.4222, dtype=torch.float64)
plot_reconstruction(dataset.unnormalise_scale(x).numpy(), dataset.index_colors, root=result_save_path)

Density Estimation¶
from pdmtut.vis import plot_density
from regilib.core.invertible_modules.bijective import AffineTransform
log_likelihood = model.log_likelihood(dataset.X)
# unnormalise the data and compute the change in density
un_normalise = AffineTransform(dataset._mean, 1/dataset._std)
data = un_normalise.forward(DynamicalState(state=dataset.X.clone().requires_grad_(True), log_prob=log_likelihood.clone()))
data_log_likelihood = data.log_prob.mean()
if result_save_path is not None:
with open(os.path.join(result_save_path, 'density.txt'), 'w') as f:
f.write(str(data_log_likelihood.item()))
data_log_likelihood
tensor(-8.5386, dtype=torch.float64)
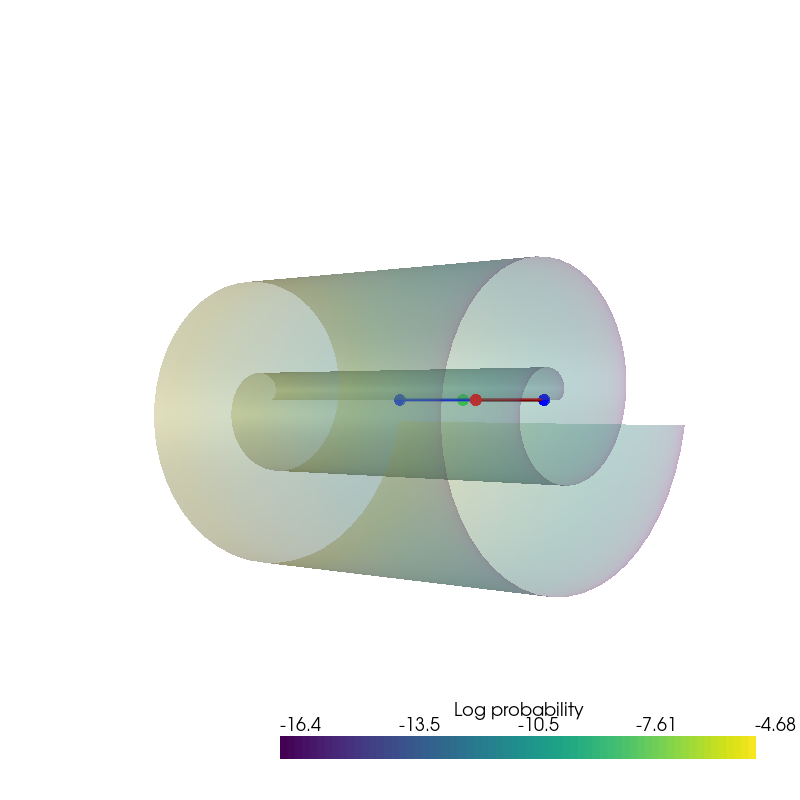
plot_density(data.state.detach().numpy(), data.log_prob.detach().numpy(), root=result_save_path)
Generate Samples¶
from pdmtut.vis import plot_generated_samples
from regilib.core.invertible_modules.bijective import AffineTransform
generated_samples = model.sample_posterior(100**2)
generated_samples_log_likelihood = model.log_likelihood(generated_samples)
# unnormalise the data and compute the change in density
un_normalise = AffineTransform(dataset._mean, 1/dataset._std)
data = un_normalise.forward(DynamicalState(state=generated_samples.clone().requires_grad_(True), log_prob=generated_samples_log_likelihood.clone()))
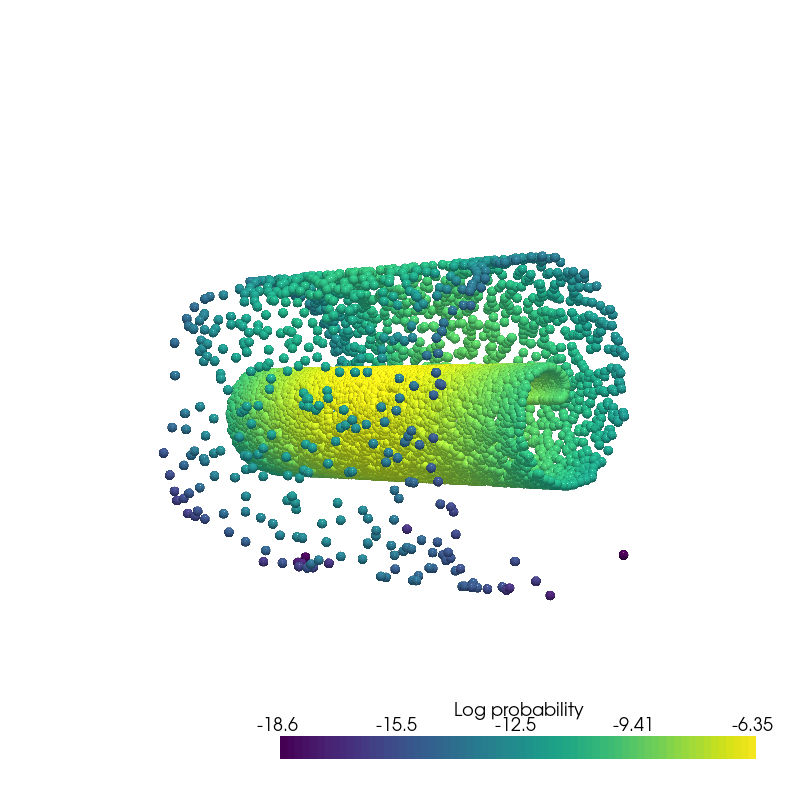
plot_generated_samples(data.state.detach().numpy(), data.log_prob.detach().numpy(), root=result_save_path)
Interpolation¶
from pdmtut.vis import plot_interpolation
from scipy.interpolate import interp1d
z_extremes = model.encode(dataset.y_extremes)
uniform_state, uniform_log_prob, _ = dataset.sample_points_uniformly(n_samples=100**2, seed=11)
linfit1 = interp1d([1,20], z_extremes[:2].numpy(), axis=0)
linfit2 = interp1d([1,20], z_extremes[2:].numpy(), axis=0)
linfit3 = interp1d([1,20], z_extremes[[1,2]].numpy(), axis=0)
interpolated_points_1 = model.decode(torch.Tensor(linfit1(np.arange(1,21))))
interpolated_points_2 = model.decode(torch.Tensor(linfit2(np.arange(1,21))))
interpolated_points_3 = model.decode(torch.Tensor(linfit3(np.arange(1,21))))
/home/bawaw/.conda/envs/pdm_tutorial/lib/python3.8/site-packages/torch/functional.py:445: UserWarning: torch.meshgrid: in an upcoming release, it will be required to pass the indexing argument. (Triggered internally at /opt/conda/conda-bld/pytorch_1639180588308/work/aten/src/ATen/native/TensorShape.cpp:2157.)
return _VF.meshgrid(tensors, **kwargs) # type: ignore[attr-defined]
plot_interpolation(
dataset.unnormalise_scale(interpolated_points_1).numpy(),
dataset.unnormalise_scale(interpolated_points_2).numpy(),
dataset.unnormalise_scale(interpolated_points_3).numpy(),
uniform_state.detach().view(100, 100, 3).permute(2, 0, 1).numpy(),
uniform_log_prob.numpy(), root=result_save_path
)

Experiment 1b: swiss roll (3-components)¶
pv.set_plot_theme("document")
model_save_path = '../results/swiss_roll/ppca_3_components'
if store_results:
result_save_path = '../results/swiss_roll/ppca_3_components'
pv.set_jupyter_backend('None')
else:
pv.set_jupyter_backend('ipygany')
result_save_path = None
dataset = SwissRoll(n_samples=100**2, seed=11)
if load_models and PPCAModel.save_exists(model_save_path):
model = PPCAModel.load(model_save_path)
else:
model = PPCAModel(n_components=3)
model.fit_model(dataset.X, result_save_path)
if store_results:
model.save(model_save_path)
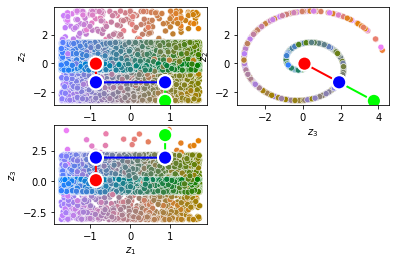
from pdmtut.vis import plot_representation
z = model.encode(dataset.X)
z_extremes = model.encode(dataset.y_extremes)
z_extremes = torch.cat([z_extremes, z_extremes[[1,2]]])
plot_representation(z.numpy(), index_colors=dataset.index_colors, z_extremes=z_extremes, interpolate_background=True, root=result_save_path)
Input Reconstruction¶
from pdmtut.vis import plot_reconstruction
z = model.encode(dataset.X)
x = model.decode(z)
mse = (dataset.unnormalise_scale(dataset.X) - dataset.unnormalise_scale(x)).pow(2).sum(-1).mean()
if result_save_path is not None:
with open(os.path.join(result_save_path, 'reconstruction.txt'), 'w') as f:
f.write(str(mse.item()))
mse
tensor(5.9800e-14, dtype=torch.float64)
plot_reconstruction(dataset.unnormalise_scale(x).numpy(), dataset.index_colors, root=result_save_path)
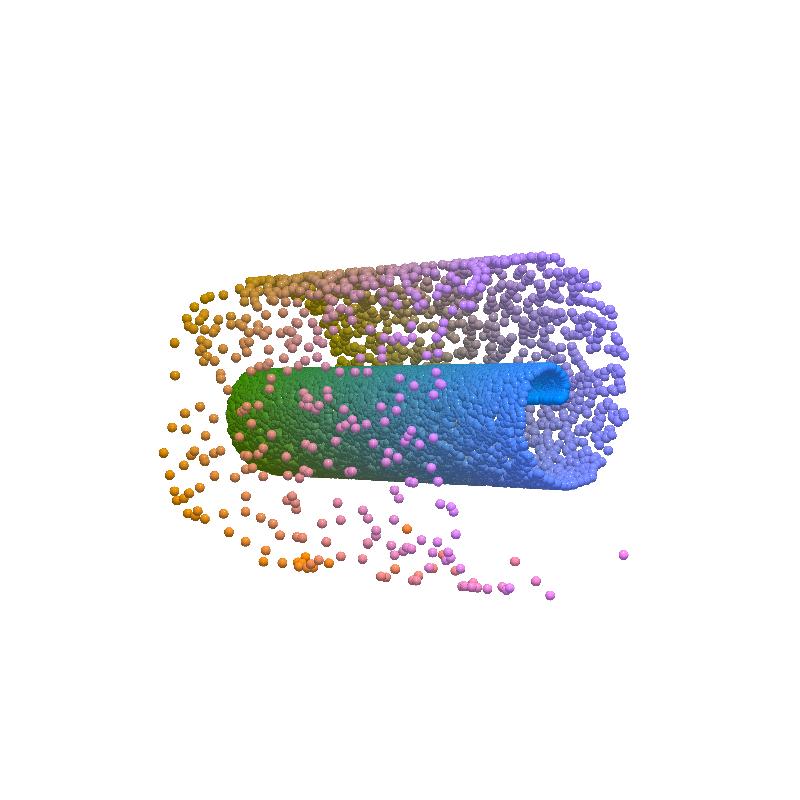
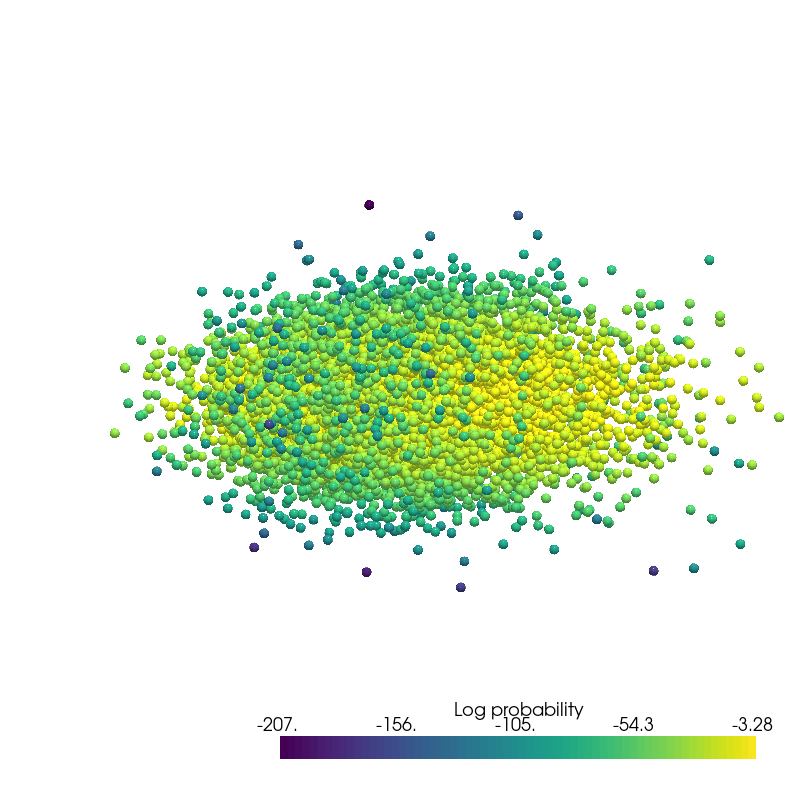
Density Estimation¶
from pdmtut.vis import plot_density
from regilib.core.invertible_modules.bijective import AffineTransform
log_likelihood = model.log_likelihood(dataset.X)
# unnormalise the data and compute the change in density
un_normalise = AffineTransform(dataset._mean, 1/dataset._std)
data = un_normalise.forward(DynamicalState(state=dataset.X.clone().requires_grad_(True), log_prob=log_likelihood.clone()))
data_log_likelihood = data.log_prob.mean()
if result_save_path is not None:
with open(os.path.join(result_save_path, 'density.txt'), 'w') as f:
f.write(str(data_log_likelihood.item()))
data_log_likelihood
tensor(-25.1159, dtype=torch.float64)
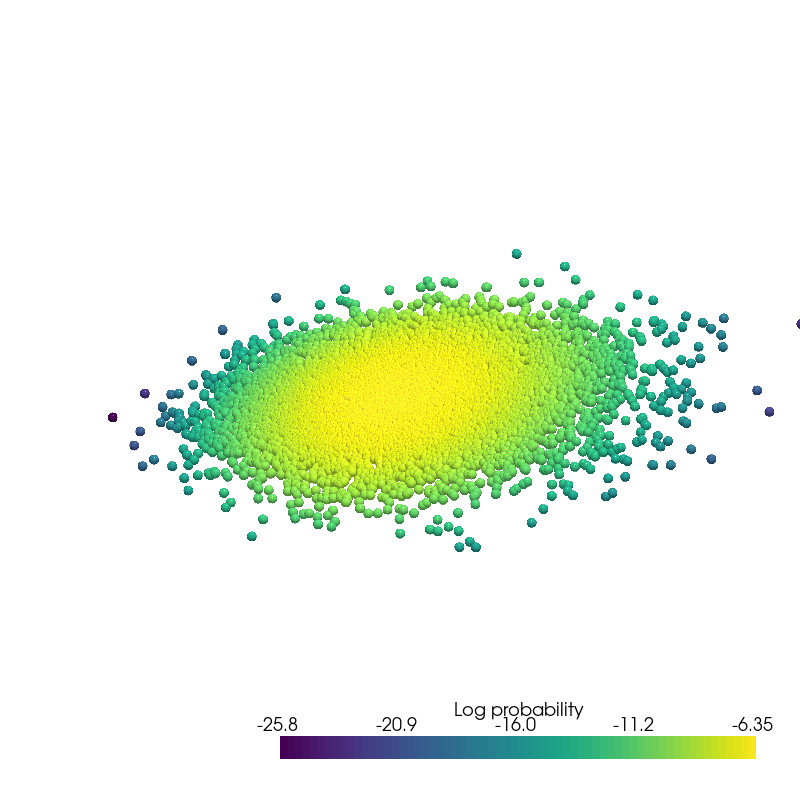
plot_density(data.state.detach().numpy(), data.log_prob.detach().numpy(), root=result_save_path)

Generate Samples¶
from pdmtut.vis import plot_generated_samples
from regilib.core.invertible_modules.bijective import AffineTransform
generated_samples = model.sample_posterior(100**2)
generated_samples_log_likelihood = model.log_likelihood(generated_samples)
# unnormalise the data and compute the change in density
un_normalise = AffineTransform(dataset._mean, 1/dataset._std)
data = un_normalise.forward(DynamicalState(state=generated_samples.clone().requires_grad_(True), log_prob=generated_samples_log_likelihood.clone()))
plot_generated_samples(data.state.detach().numpy(), data.log_prob.detach().numpy(), root=result_save_path)
Interpolation¶
from pdmtut.vis import plot_interpolation
from scipy.interpolate import interp1d
z_extremes = model.encode(dataset.y_extremes)
uniform_state, uniform_log_prob, _ = dataset.sample_points_uniformly(n_samples=100**2, seed=11)
linfit1 = interp1d([1,20], z_extremes[:2].numpy(), axis=0)
linfit2 = interp1d([1,20], z_extremes[2:].numpy(), axis=0)
linfit3 = interp1d([1,20], z_extremes[[1,2]].numpy(), axis=0)
interpolated_points_1 = model.decode(torch.Tensor(linfit1(np.arange(1,21))))
interpolated_points_2 = model.decode(torch.Tensor(linfit2(np.arange(1,21))))
interpolated_points_3 = model.decode(torch.Tensor(linfit3(np.arange(1,21))))
plot_interpolation(
dataset.unnormalise_scale(interpolated_points_1).numpy(),
dataset.unnormalise_scale(interpolated_points_2).numpy(),
dataset.unnormalise_scale(interpolated_points_3).numpy(),
uniform_state.detach().view(100, 100, 3).permute(2, 0, 1).numpy(),
uniform_log_prob.numpy(), root=result_save_path
)